- nodes = pixels
- edges = between pairsof pixels
- affinity weight, measuring similarity, for each edge
- similarity is inversely proportional to difference
goal
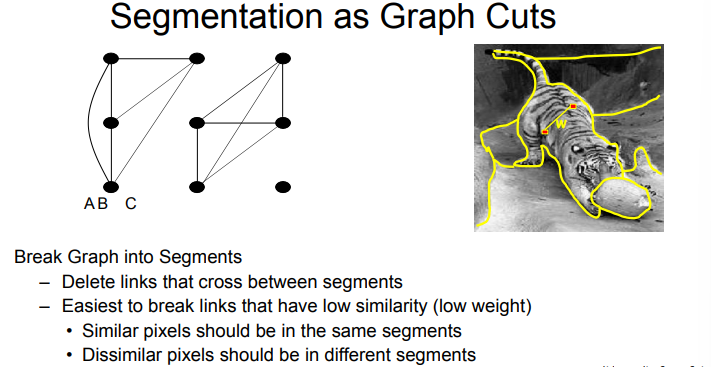
- cut graph into segments where
- pixels in segments are highly similar
- pixels across segments are dissimilar
- construct graph - what edges?
- fully connected - all pairwise sims, but infeasible
- grid-based (neighboring pixels) -
- more efficient than fully connected graph
- only gets local interactions
- local neighborhood
- reasonably fast, graph still sparse
- good tradeoff
- define similarity
- gaussian similarity
- solve for e-vecs, eigenvectors+values
- formulate as graph laplacian problem
- eigenvectors give cluster assignments
segmentation as graph cuts
image as graph
- Every pixel is connected to its 8 neighboring pixels
- The edges between neighbors have weights that are determined by the distance between them.
- Edge weights between pixels are determined using dist(x, x’) distance in feature space.
- where x and x’ are two neighboring pixel
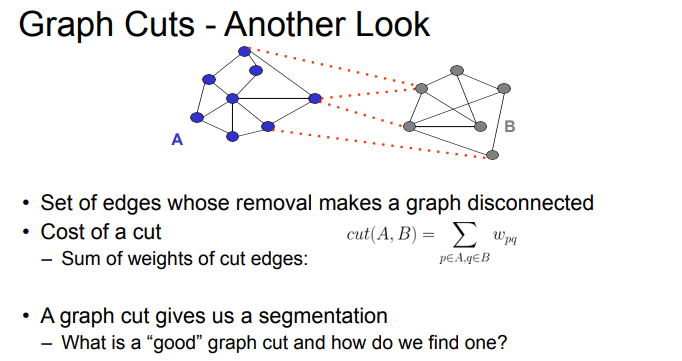
segmentation as graph-cut
- S is segmentation of G where we keep all vertices but select subset of edges
- S divides G such that G’ contains distinct clusters
alternative
clustering via eigenvalue
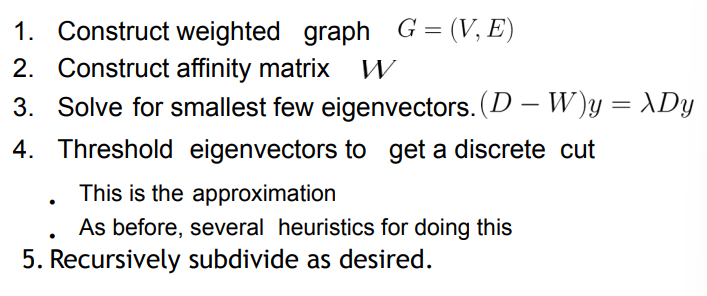
- Construct affinity matrix W
- Compute eigenvalues and vectors of W
- Until done
- Take eigenvector of largest unprocessed eigenvalue
- Zero all components of elements that have already been clustered
- Threshold remaining components to determine cluster membership Note: This is an example of a spectral clustering algorithm
min cut
- smallest num of elements (unweighted) or smallest sum of weights (weighted) drawback
- weight of cut proportional to num edges
- biased towards cutting small, isolated components
better: normalize cuts
- avoid bias towards small clusters
- balances within-cluster sim and between-cluster dissim
- normalize segment size to fix min cut’s bias
pros
- elegant
- flexible to choice of affinity matrix
cons
- computationally heavy w many cuts
- bias towards balanced partitions
- constrained by affinity matrix model